Ι. Development of techniques to study chromatin conformation
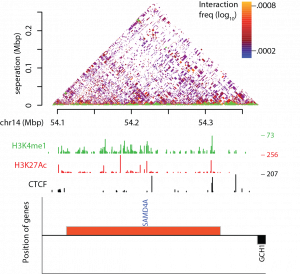
Example of T2C (adapted and modified from Kolovos et al., 2018)
Development of techniques which study the spatiotemporal chromatin architecture in various biological systems. Techniques such as 3C-seq (Stadhouders, et al., 2013) and T2C (Kolovos et al., 2014; Kolovos et al., 2018), unveil the chromatin organization
T2C (Kolovos et al., 2014; Kolovos et al., 2018) allows to study the spatiotemporal chromatin organization, the compartmentalization and the interactome at high resolution (0.4 kbp), high coverage, without the need to bin the reads and with low cost.
ΙI. Chromatin architecture and the bimodal role of Transcription Factors
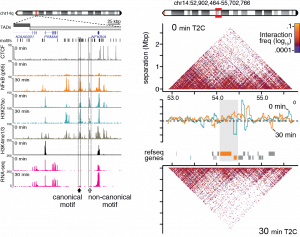
(Adapted and modified from Kolovos et al., 2016)
The study of the dynamics and of role of the transcription factors and the role of chromatin architecture in biological systems such as the hematopoietic differentiation (Kolovos et al., in preparation) and inflammation response of endothelial cells (Diermeier, et al. 2014; Kolovos et al., 2016). These revealed a combinatorial network of transcription factors which control gene regulation and expression via predetermined chromatin interactions (looping)
ΙIΙ. Regulation of gene expression in response to signalling pathways
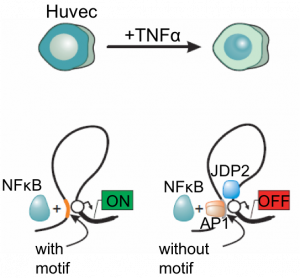
Bimobal role of NFκB in HUVECs (results concern Kolovos et al., 2016)
The study and the analysis of the dynamics of enhancers/promoters (poised/active/silenced) and of the 3D chromatin organization in response to signalling pathways such as TNFα και TGF, through various transcription factors active in these pathways and their role in regulation of gene expression.
VΙ. Epigenetics in relation to regulation of gene expression
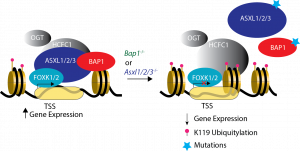
(Adapted and modified from Kolovos et al., 2020)
The polycomb proteins (PRC1/2) are important for the proper regulation of gene expression. We study the relationship of Polycomb repressive de-ubiquitinase (PR-DUB) with PRC1/2 and its role in the maintenance of chromatin in an optimal state, through epigenetic mechanism, in order to control the proper gene expression of genes important various functions of the organisms (Kolovos et al., 2020).
V. Identification of new Transcription Factors
Screening approaches with state-of-the-art techniques (CRISPR-Cas9) in order to identify new transcription factors and their role in hematopoiesis. The aim is to unveil the underlying mechanisms of hematopoietic differentiation in order to understand the molecular backbone of hematopoiesis.
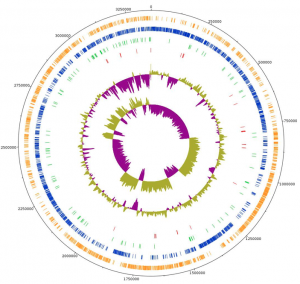
Circular genome map of L. plantarum L125 (Adapted from Tegopoulos et al., 2021)
VI. Analysis of microbiome and bacteria genome
Developing methodologies to analyze the microbiome and bacteria genome, in order to in silico taxonomically and phylogenetically clasify them, as well as to in silico analyze their genome predicting their potential probiotic, health or other properties resulting in an easier and more targeted in vitro verification (Stergiou et al., 2021, Tegopoulos et al., 2021).
